AnnoUpsetPlot
AnnoUpsetPlot.RdAnnoUpsetPlot
Arguments
- LinkageObject
An LinkageObject after the annottation.
- order_by
How the intersections in the matrix should be ordered by. Options include frequency (entered as "freq"), degree, or both in any order.
Examples
library(Linkage)
data("SmallLinkageObject")
gene_list <- c("TSPAN6", "CD99", "KLHL13")
LinkageObject <-
RegulatoryPeak(
LinkageObject = SmallLinkageObject,
gene_list = gene_list,
genelist_idtype = "external_gene_name"
)
peakAnno <- PeakAnnotation(LinkageObject, Species = "Homo")
#> Loading required package: org.Hs.eg.db
#> Loading required package: AnnotationDbi
#> Loading required package: stats4
#> Loading required package: BiocGenerics
#>
#> Attaching package: 'BiocGenerics'
#> The following objects are masked from 'package:stats':
#>
#> IQR, mad, sd, var, xtabs
#> The following objects are masked from 'package:base':
#>
#> Filter, Find, Map, Position, Reduce, anyDuplicated, aperm, append,
#> as.data.frame, basename, cbind, colnames, dirname, do.call,
#> duplicated, eval, evalq, get, grep, grepl, intersect, is.unsorted,
#> lapply, mapply, match, mget, order, paste, pmax, pmax.int, pmin,
#> pmin.int, rank, rbind, rownames, sapply, setdiff, sort, table,
#> tapply, union, unique, unsplit, which.max, which.min
#> Loading required package: Biobase
#> Welcome to Bioconductor
#>
#> Vignettes contain introductory material; view with
#> 'browseVignettes()'. To cite Bioconductor, see
#> 'citation("Biobase")', and for packages 'citation("pkgname")'.
#> Loading required package: IRanges
#> Loading required package: S4Vectors
#>
#> Attaching package: 'S4Vectors'
#> The following object is masked from 'package:utils':
#>
#> findMatches
#> The following objects are masked from 'package:base':
#>
#> I, expand.grid, unname
#>
#> Attaching package: 'IRanges'
#> The following object is masked from 'package:grDevices':
#>
#> windows
#>
#> 'select()' returned 1:1 mapping between keys and columns
#> 'select()' returned 1:many mapping between keys and columns
#> 'select()' returned 1:1 mapping between keys and columns
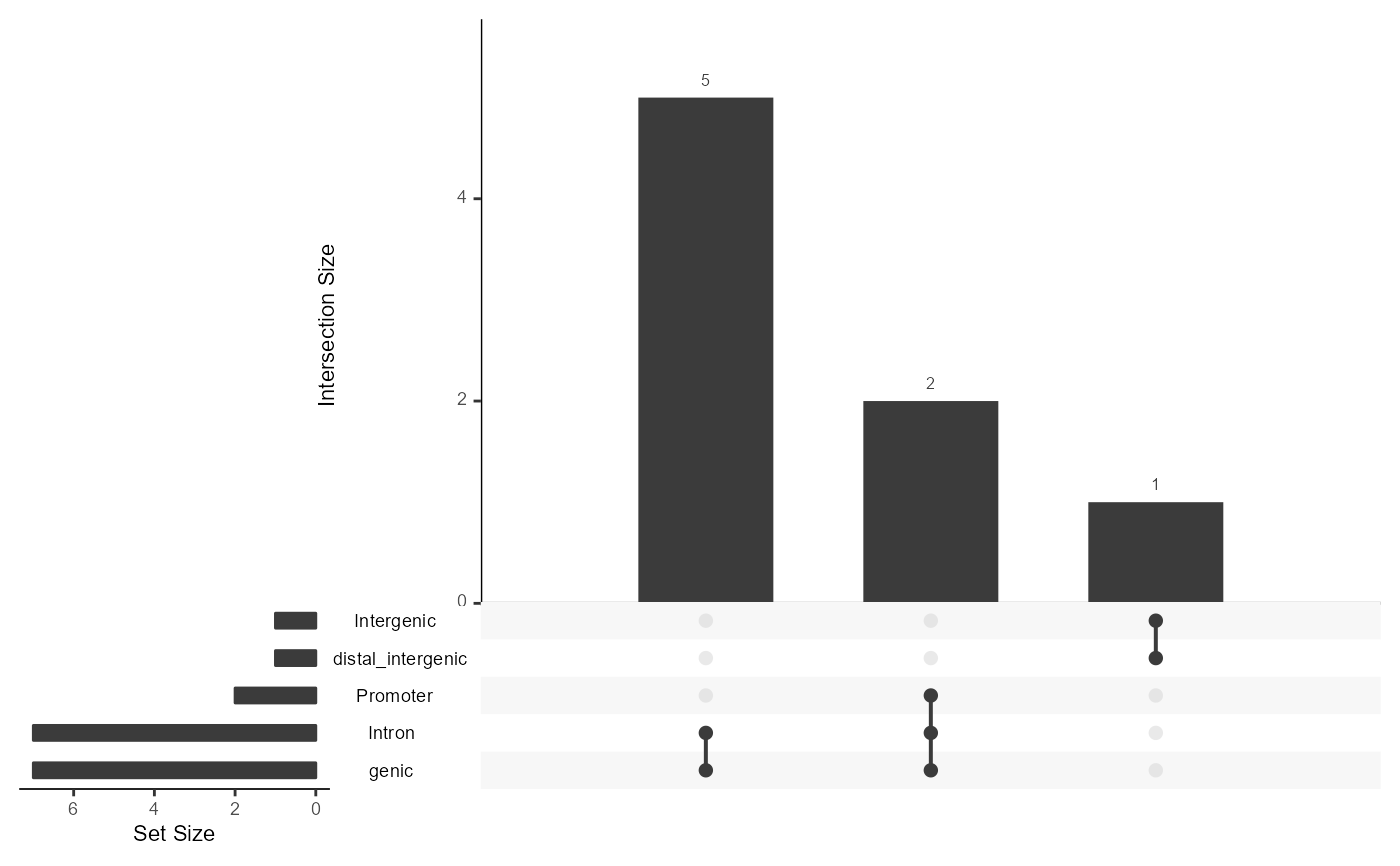
AnnoUpsetPlot(LinkageObject = peakAnno)
#> Warning: `cols` is now required when using `unnest()`.
#> ℹ Please use `cols = c(res)`.